GSoC
Work continues on this front.
Both Aditya and Ioannis already have pull requests open, so I've been reviewing them and providing feedback.
We're making sure we follow best practices right from the start---so doc strings, unit tests, type hints are all already in use.
I also added coverage information to our pyNeuroML CI.
It doesn't use a third party service like Coveralls/Codecov/Codespace at the moment.
It's a combination of two GitHub actions---one summarises the coverage that we generate using pytest-cov, and the other comments on the pull request.
It works well enough.
The commenter action replaces to same comment each time too, so it doesn't spam the pull request with repeated comments.
This has pointed out that some of our code needs more testing, so we've put that on our agenda too now.
Model conversion
I continued to work on converting the Purkinje cell model from Zang et al.
Using my test script, I've converted another couple of channels.
While converting the "Large conductance Ca2+ activated K+ channel" (mslo.mod), I was reminded of this excellent features of LEMS:
- LEMS is unit and dimension aware: one does not have to worry about units while writing equations, one can use whatever units one wishes and LEMS will convert them to the right multipliers depending on the dimensions.
So, for example, even if a model component provides the temperature value in degrees C, LEMS will convert this into Kelvin when it's being used somewhere.
Here's how it needs to be explicitly converted in the mod file:
alpha = exp(Qo*FARADAY*v/R/(273.15 + celsius))
Here's how the corresponding LEMS looks:
<DerivedVariable name="alpha_beta" dimension="none" value="exp((Qo_Qc * Faraday * v/R) / EXP_TEMP_CELSIUS)"/>
(The variable names have been tweaked so that the same derived variable can also be used for other calculations that follow the same formalism)
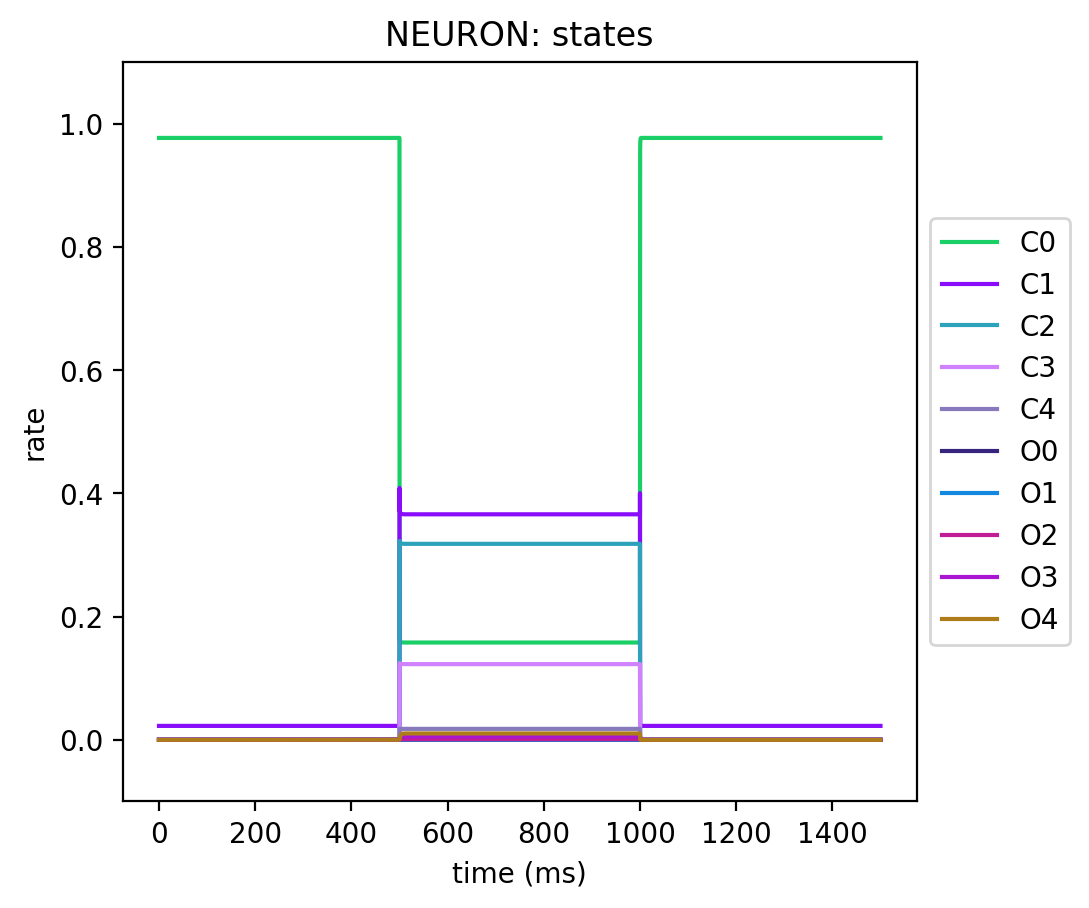
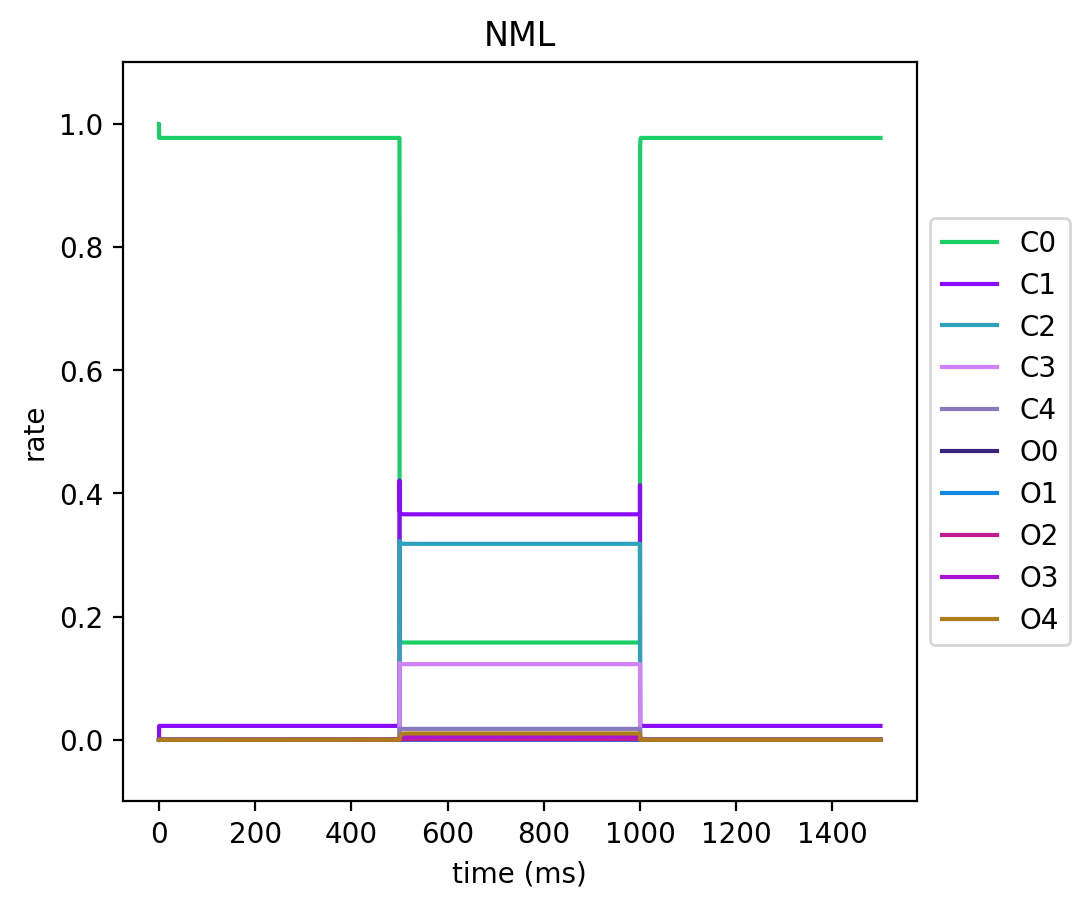
Here are the plots for the kinetic state variables for the NEURON and NeuroML conversions for a simple protocol where we:
- create a simple single compartmental cell, with only a soma
- add the ion channel to it
- add the minimal biophysics
- use a "Calcium clamp" to set the intra- and extra-cellular calcium concentrations to some values for a part of the simulation
We then record all the states and plot them to see how our conversion went.
You'll see that they match almost exactly, which is always a good sign.
There are two more channels left to convert.
These are of a third type---they use the "GHK" (Goldman-Hodgkin-Katz) equation.
I'll work on converting them next.
Then, we need to place these on the cell morphology to verify that the NEURON implementation and our standardised NeuroML implementation show the same dynamics.

Comments