Week 22 update
Last Monday was the late spring bank holiday, so it was a four day week.
NeuroML related

NeuroML is a standard and software ecosystem for biophysically detailed neuronal modelling.
We've made some changes to the NeuroML standard in the last week. For neuron models, a few basic properties were optional so far:
This is because the NEURON simulator, which is sort of the reference simulator for computational neuro-modelling, would use sane defaults if these values were not provided by the user. However, since other simulators that NeuroML supports may not fill in defaults, we decided that these properties should also be compulsory for the user to provide explicitly.
Updating the standard requires a few steps:
- update the XSD schema
- regenerate the various APIs, e.g., libNeuroML
- run tests on all our models to see how many break with this backwards-incompatible change and fix them
The fall out doesn't seem too bad, and we've fixed the few models that needed fixes.
Toggling build time validation in libNeuroML
One of the strengths of the NeuroML standard is that we have a schema that allows the validation of NeuroML model descriptions---before the models have been initialised/instantiated/simulated.
Using our factory functions, the model is continuously validated at each construction/build step.
While this is useful, it can also be a hindrance to users in some cases.
For example, certain model components, in general usage, will not be valid when initially created.
A Network component is one such example.
It must contain at least one Population.
However, in normal usage, one generally creates a network and then adds populations to it.
So, when the network is initially created, it will be invalid.
In libNeuroML, validation errors will throw a ValueError immediately to inform the user, and halting the Python script.
There are other such cases too, and we thought this conflicts with our goal of making it easier for users to create NeuroML models.
A middle-ground that we've come up with is the ability to globally toggle the state of build time validation.
By default, it's enabled, but users may disable it if required---explicitly.
Additionally, build time validation can also be disabled for the specific call to the factory function using validate=False.
The latter is recommended.
When build time validation is disabled, it will throw a user warning instead of the ValueError.
Completing the spike plotting bits
Aditya was working on refactoring the pyNeuroML code that plots spikes from simulation output.
He completed his PR and I merged it after making a few tweaks to the tests.
One can now easily plot spikes from data files using the pynml-plotspikes command, and also using the related functions in the pyNeuroML API.
Here's an example of plotting spikes from Brunel et al (2000).
$ pynml-plotspikes -format t_id *_net.spikes
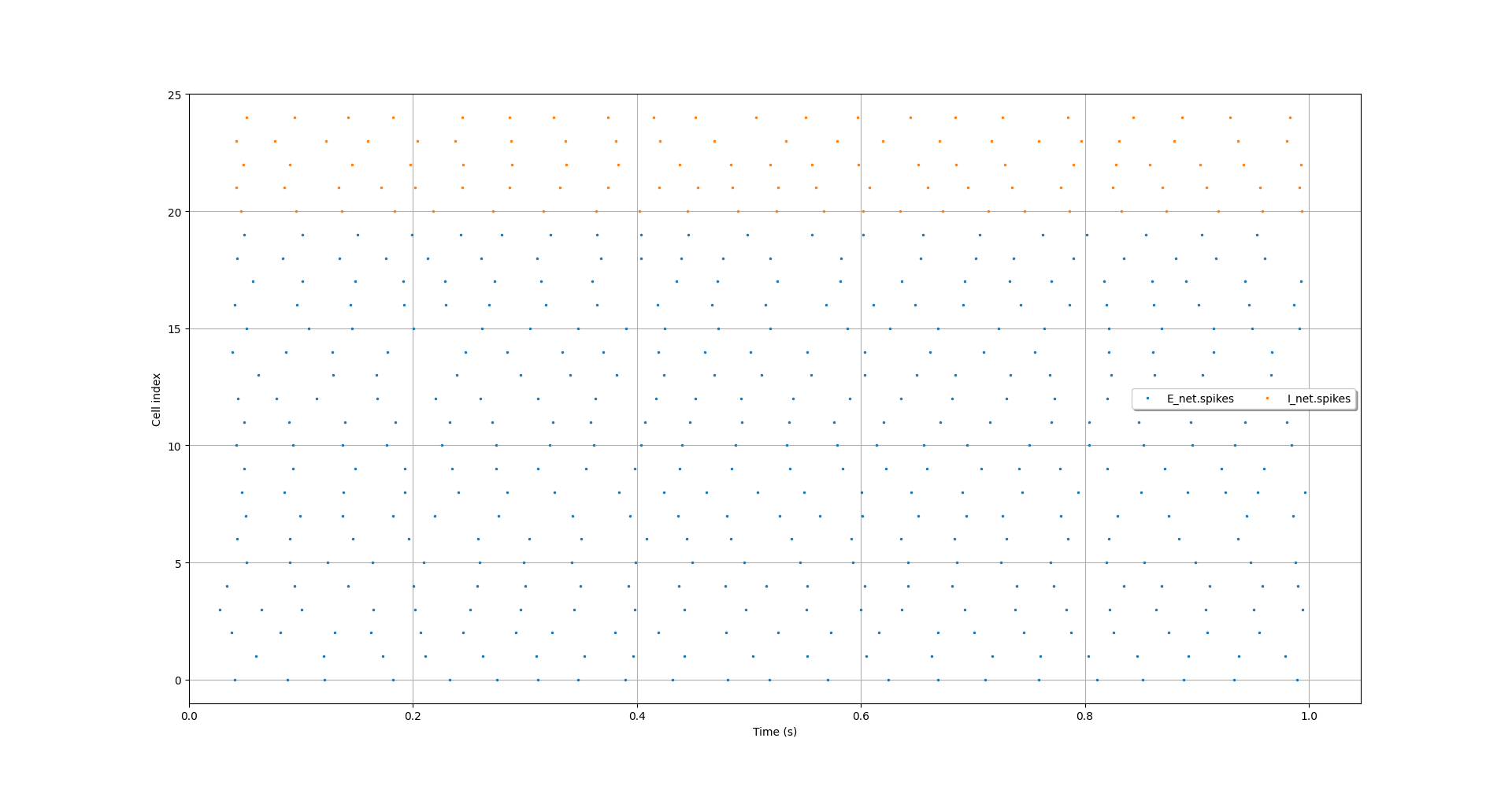
Raster plot showing spikes from a simulation of Brunel et al (2000).
GSoC
We had our first meeting of the contribution period last week. Both candidates, Aditya and Ioannis, are already set up and well on their way. Ioannis is continuing with the task that he had begun working on during the contribution period already---trying to get an "upright" view of single neurons in our 3D visualiser. Aditya has started working on loading SWC files of neuronal reconstructions into an appropriate data structure that will make it easy to query the SWC morphology and validate it, and then eventually convert it into NeuroML.
Other stuff
I continued to work on converting the Purkinje cell model from Zang et al. I now have a test scripts that plots the various state variables from the ion channels represented using kinetic schemes. It adds the ion channel to a single compartmental cell, gives it a constant current to bring the state variables to their steady states, and plots these. It does this both for the NEURON implementation and the NeuroML one so that I can compare the dynamics.
Fedora related
Fixes, updates, and bug filing continues in the NeuroFedora SIG. It's ticking along nicely, as it tends to do.
Comments