Testing out Sumatra: a tool for managing iterations of simulations/analyses
In the ~4 years that I've spent on my PhD now, I've run hundreds, nay, thousands of simulations. Research work is incredibly iterative. I (and I assume others too) make small changes to their methods and then study how these changes produce different results, and this cycle continues until a proposed hypothesis has either been accepted or refuted (or a completely new insight gained, which happens quite often too!).
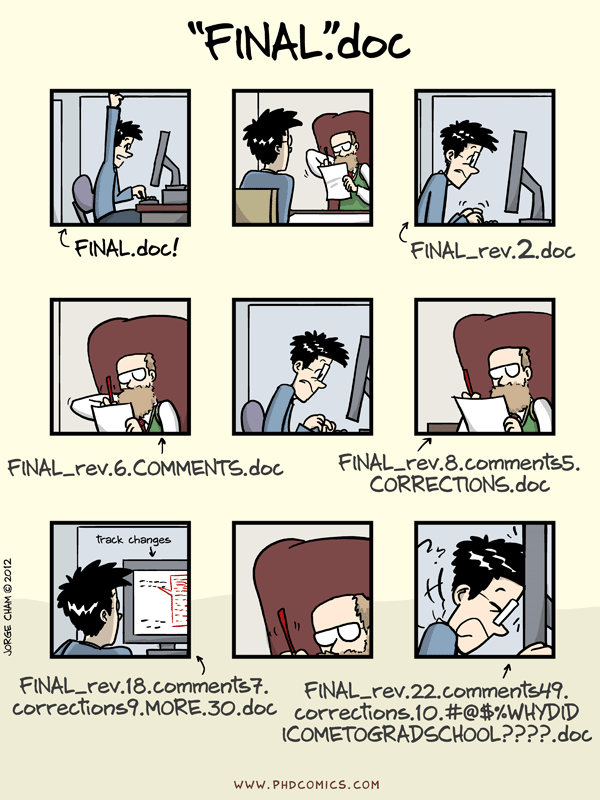
Folders, and Dropbox? Please, no.
Keeping track of all these iterations is quite a task. I've seen multiple methods that people use to do this. A popular method is to make a different folder for each different version of code, and then use something like Dropbox to store them all.
Since I come from a computing background, I firmly believe that this is not a good way of going about it. It may work for folks---people I know and work with use this method---but it is simply a bad way of going about it. This PhDComic does a rather good job of showing an example situation. Sure, this is about a document, but when source code is kept in different folders, a similar situation arises. You get the idea.
Version control, YES!
If there weren't tools designed to track and manage such projects, one could still argue for using such methods, but the truth is that there is a plethora of version control tools available under Free/Open Source licenses. Not only do these tools manage projects, they also make collaborating over source code simple.
All my simulation code, for example, lives in a Git repository (which will be made available under a Free/Open source license as soon as my paper goes out to ensure that others can read, verify, and build on it). The support scripts that I use to set up simulations and then analyse the data they produce already live here on GitHub, for example. Please go ahead and use them if they fit your purpose.
I have different Git branches for different features that I add to the simulations---the different hypothesis that I'm testing out. I also keep a rather meticulous record of everything I do in a research journal in LaTeX that also lives in a Git repository, and uses Calliope (a simple helper script to manage various journaling tasks). Everything goes in here---graphs, images, sometimes patches and source code even, and the deductions and other comments/thoughts too.
My rather simple system is as follows:
- Each new feature/hypothesis gets its own Git branch.
- Each version of its implementation, therefore, gets its own unique commit (a snapshot of code that Git saves for the user with a unique identifier and a complete record of the changes that were made to the project, when they were made and so on.)
- For each run of a snapshot, the generated data is stored in a folder that is named YYYYMMDDHHMM (Year, month, day, time), which, unless you figure out how to go back in time, is also unique.
- The commit hash + YYYYMMDD become a unique identifier for each code snapshot and the results that it generated.
- A new chapter in my research journal holds a summary of the simulation, and all the analysis that I do. I even name the chapter "git-hash/YYYYMMDDHHMM".

I know that learning a version control system has a steep initial curve, but I really do think that this is one tool that is well worth the time.
Using a version control system has many advantages, some of which are:
- It lets you keep the full history of your source code, and go back to any previous version.
- You know exactly what you changed between two snapshots.
- If multiple people work on the code, everyone knows exactly who authored what.
- These tools make changing code, trying out things, and so on, very very easy. Try something out in a different branch, if it worked, yay, keep the branch running; maybe even merge it to the main branch? If it didn't make a note, delete the branch, and move on!
- With services like GitHub, BitBucket, and GitLab, collaboration becomes really easy.
- Ah, and note, that every collaborator has a copy of the source code, so it has been backed up too! Even if you work alone, there's always another copy on GitHub (or whatever service you use).
Here's a quick beginners guide to using Git and GitHub: http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004668 There are many more all over the WWW, of course. Duckduckgo is your friend. (Why Duckduckgo and not Google?)
What's Sumatra about, then?
I've been meaning to try Sumatra out for a while now. What Sumatra does is sort of bring the functions of all my scripts together into one well-designed tool. Sumatra can do the running bit, then save the generated data in a unique location, and it even lets users add comments about the simulation. Sumatra even has a web based front end for those that would prefer a graphical interface instead of the command line. Lastly, Sumatra is written in Python, so it works on pretty much all systems. Note that Sumatra forces the use of a version control system (from what I've seen yet).
A quick walk-through
The documentation contains all of this already, but I'll show the steps here too. I used a dummy repository to test it out.
Installing Sumatra is as easy as a pip command. I would suggest setting up a virtual-environment, though:
python3 -m venv --system-site-packages sumatra-virtual
We then activate the virtual-environment, and install Sumatra:
source sumatra-virtual/bin/activate pip install sumatra
Once it finishes installing, simply mark a version controlled source repository as managed by Sumatra:
cd my-awesome-project smt init my-awesome-project
Then, one can see the information that Sumatra has on the project, for example:
smt info Project name : test-repo Default executable : Python (version: 3.6.5) at /home/asinha/dump/sumatra-virt/bin/python3 Default repository : GitRepository at /home/asinha/Documents/02_Code/00_repos/00_mine/sumatra-nest-cluster-test (upstream: git@github.com:sanjayankur31/sumatra-nest-cluster-test.git) Default main file : test.py Default launch mode : serial Data store (output) : /home/asinha/Documents/02_Code/00_repos/00_mine/sumatra-nest-cluster-test/Data . (input) : / Record store : Django (/home/asinha/Documents/02_Code/00_repos/00_mine/sumatra-nest-cluster-test/.smt/records) Code change policy : error Append label to : None Label generator : timestamp Timestamp format : %Y%m%d-%H%M%S Plug-ins : [] Sumatra version : 0.7.4
My test script only prints a short message. Here's how one would run it using Sumatra:
# so that we don't have to specify this for each run smt configure --executable=python3 --main=test.py smt run Hello Sumatra World! Record label for this run: '20180512-200859' No data produced.
One can now see all the runs of this simulation that have been made!
smt list --long
--------------------------------------------------------------------------------
Label : 20180512-200859
Timestamp : 2018-05-12 20:08:59.761849
Reason :
Outcome :
Duration : 0.050611019134521484
Repository : GitRepository at /home/asinha/Documents/02_Code/00_repos/00_mine/sumatra-nest-
: cluster-test (upstream: git@github.com:sanjayankur31/sumatra-nest-cluster-
: test.git)
Main_File : test.py
Version : 6f4e1bf05f223a0100ca6f843c11ef4fd70490f3
Script_Arguments :
Executable : Python (version: 3.6.5) at /home/asinha/dump/sumatra-virt/bin/python3
Parameters :
Input_Data : []
Launch_Mode : serial
Output_Data : []
User : Ankur Sinha (Ankur Sinha Gmail) <sanjay.ankur@gmail.com>
Tags :
Repeats : None
--------------------------------------------------------------------------------
Label : 20180512-181422
Timestamp : 2018-05-12 18:14:22.668655
Reason :
Outcome : Well that worked
Duration : 0.05211901664733887
Repository : GitRepository at /home/asinha/Documents/02_Code/00_repos/00_mine/sumatra-nest-
: cluster-test (upstream: git@github.com:sanjayankur31/sumatra-nest-cluster-
: test.git)
Main_File : test.py
Version : 4f151a368b1fee1fa8f21026c3b6d2c6b2531da8
Script_Arguments :
Executable : Python (version: 3.6.5) at /home/asinha/dump/sumatra-virt/bin/python3
Parameters :
Input_Data : []
Launch_Mode : serial
Output_Data : []
User : Ankur Sinha (Ankur Sinha Gmail) <sanjay.ankur@gmail.com>
Tags :
Repeats : None
There's a lot more that can be done, of course. I'll quickly show the GUI version here.
One can run the webversion using:
smtweb -p 8001 #whatever port number one wants to use
Then, it'll open up in your default web-browser at http://127.0.0.1:8001/.
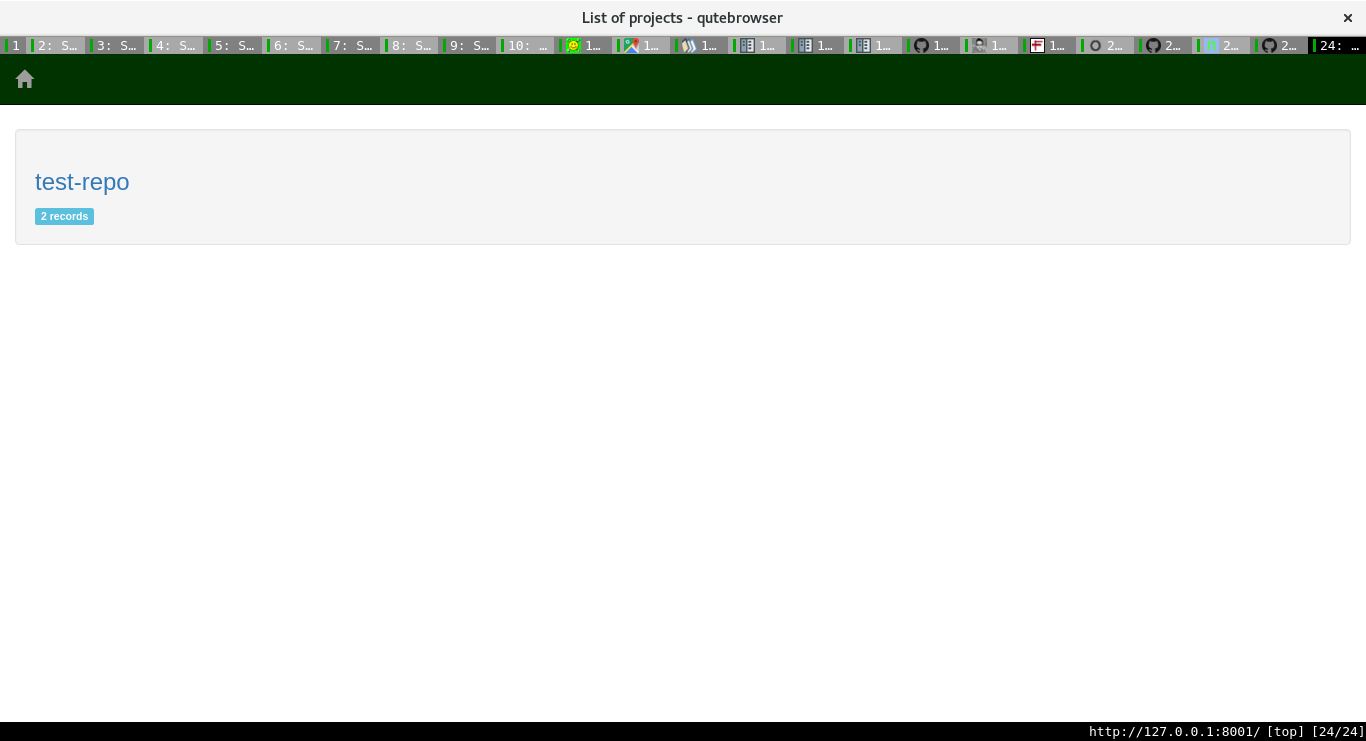
For each project, one can see the various runs, with all the associated information too.
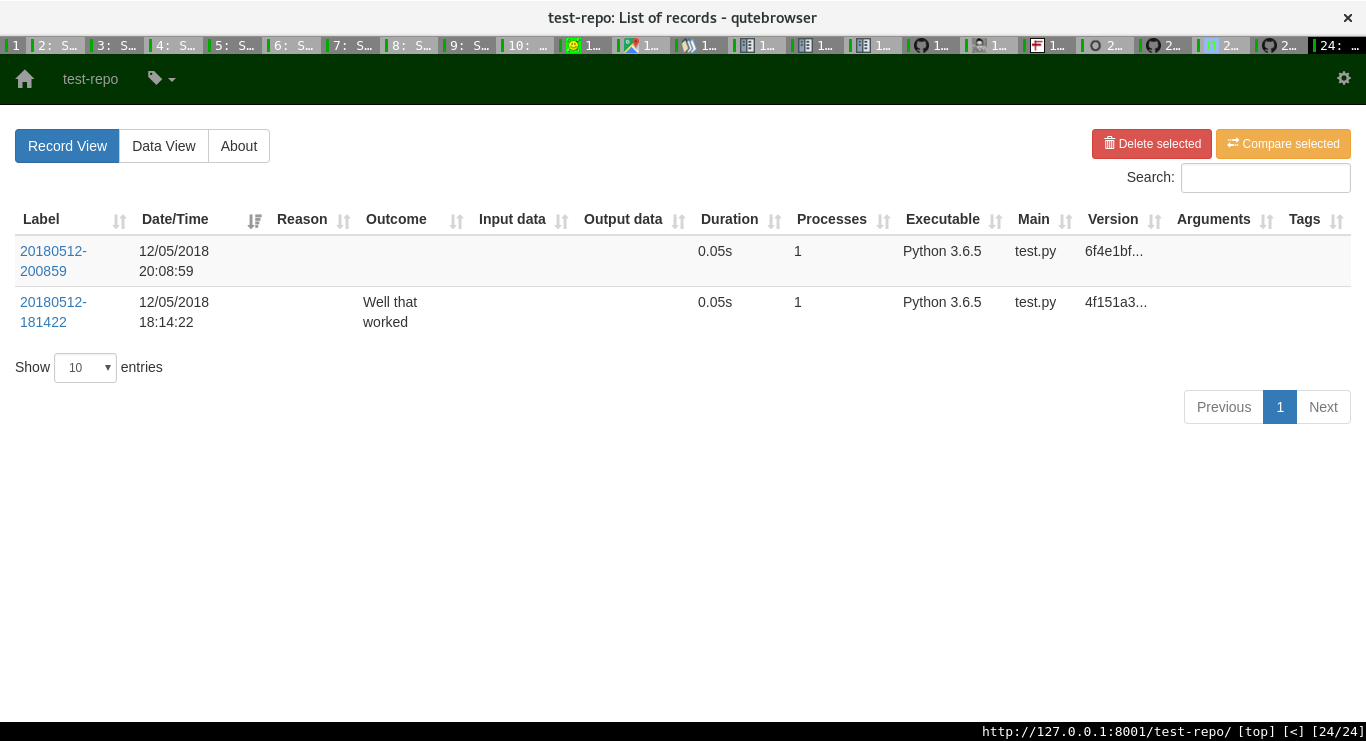
One can then add more information about a run. Sumatra already stores lots of important information as the image shows:
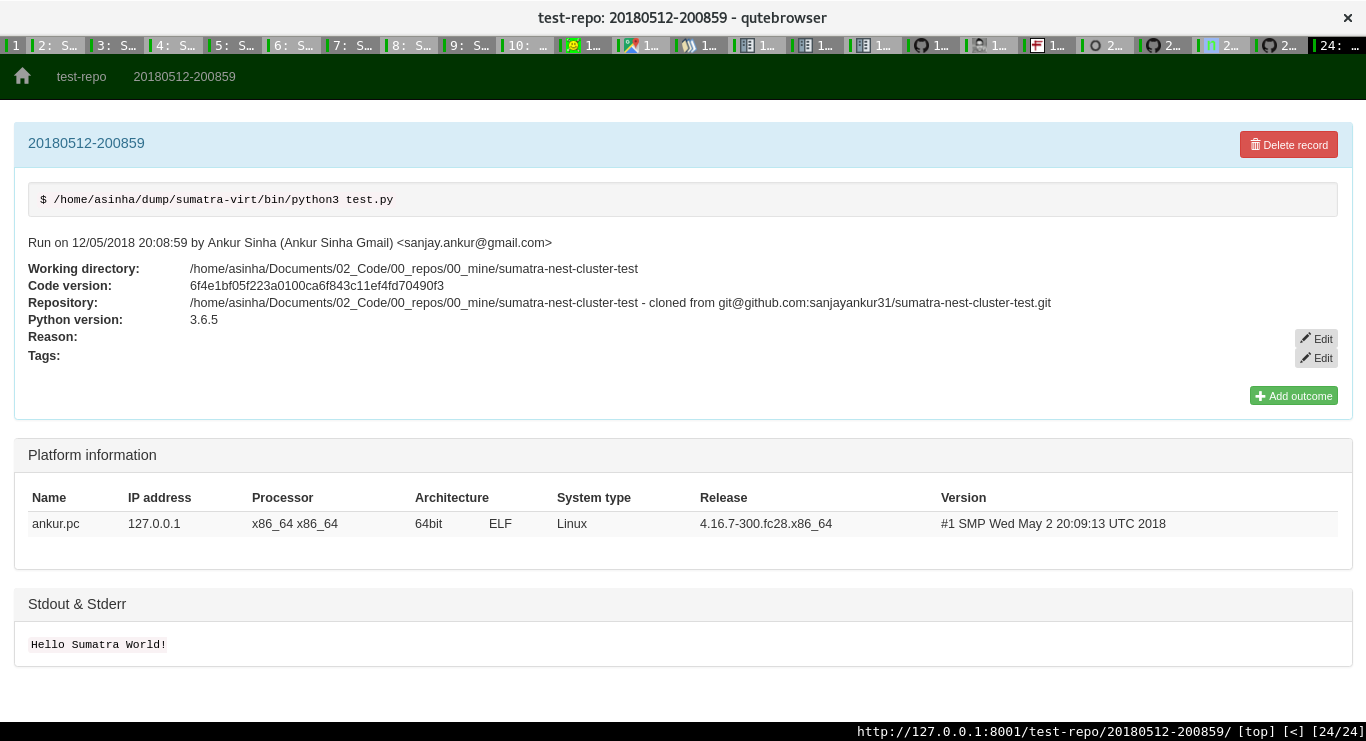
Pretty neat, huh?
I run my simulations on a cluster, and so have my own system to submit jobs to the queue system. Sumatra can run jobs in parallel on a cluster, but I've still got to check if it also integrates with the queue system that our cluster runs. Luckily, Sumatra also provides an API, so I should be able to write a few Python scripts to handle that bit too. It's on my TODO list now.
Please use version control and a Sumatra style record keeper
I haven't found another tool that does what Sumatra does yet. Maybe Jupyter notebooks would come close, but one would have to add some sort of wrapper around them to keep proper records. It'll probably be similar to my current system.
In summary, please use version control, and use a record keeper to manage and track simulations. Not only does it make it easier for you, the researcher, it also makes it easier for others to replicate the simulation since the record keeper provides all the information required to re-run the simulation.
Free/Open source software promotes Open Science
(The original video is at the Free Software Foundation's website.)
As a concluding plea, I request everyone to please use Free/Open source software for all research. Not only are these available free of cost, they provide everyone with the right to read, validate, study, copy, share, and modify the software. One can learn so much from reading how research tools are built. One can be absolutely sure of their results if they can see the code that carries out the analysis. One can build on others' work if the source is available for all to use and change. How easy does replication become when the source and all related resources are given out for all to use?
Do not use Microsoft Word, for example. Not everyone, even today, has access to Microsoft software. Should researchers be required to buy a Microsoft license to be able to collaborate with us? The tools are here to enable science, not hamper it. Proprietary software and formats do not enable science, they restrict it to those that can pay for such software. This is not a restriction we should endorse in any way.
Yes, I know that sometimes there aren't Free/Open source software alternatives that carry the same set of features, but a little bit of extra work, for me, is an investment towards Open Science. Instead of Word, as an example, use Libreoffice, or LaTeX. Use Open formats. There will be bugs, but until we report them, they will not be fixed. Until these Free/Open source tools replace restricted software as the standard for science, they will only have small communities around them that build and maintain them.
Open Science is a necessity. Researchers from the neuroscience community recently signed this letter committing to the use of Free/Open source software for their research. There are similar initiatives in other fields too, and of course, one must be aware of the Open Access movement etc.
I've made this plea in the context of science, but the video should also show you how in everyday life, it is important to use Free/Open source resources. Please use Free/Open source resources, as much as possible.
Comments